animate¶
Create movies (animations) by stitching together figures created by plotting multiple datasets, typically originating from a data load with wildcard/regex.
Saving the animation to a file requires an ffmpeg installation.
Command line¶
Command help
pgkyl animate -h
Usage: pgkyl anim [OPTIONS]
Animate the actively loaded dataset and show resulting plots in a loop.
Typically, the datasets are loaded using wildcard/regex feature of the -f
option to the main pgkyl executable. To save the animation ffmpeg needs to
be installed.
Options:
-u, --use TEXT Specify a tag to plot.
-s, --squeeze Squeeze the components into one panel.
-b, --subplots Make subplots from multiple datasets.
--nsubplotrow INTEGER Manually set the number of rows for subplots.
--nsubplotcol INTEGER Manually set the number of columns for subplots.
--transpose Transpose axes.
-c, --contour Make contour plot.
-q, --quiver Make quiver plot.
-l, --streamline Make streamline plot.
-g, --group [0|1] Switch to group mode.
-s, --scatter Make scatter plot.
--markersize FLOAT Set marker size for scatter plots.
--style TEXT Specify Matplotlib style file (default:
Postgkyl).
-d, --diverging Switch to diverging colormesh mode.
--arg TEXT Additional plotting arguments, e.g., '*--'.
-a, --fix-aspect Enforce the same scaling on both axes.
--logx Set x-axis to log scale.
--logy Set y-axis to log scale.
--logz Set values of 2D plot to log scale.
--xscale FLOAT Value to scale the x-axis (default: 1.0).
--yscale FLOAT Value to scale the y-axis (default: 1.0).
--vmax FLOAT Set maximal value of data for plots.
--vmin FLOAT Set minimal value of data for plots.
-f, --float Choose min/max levels based on current frame
(i.e., each frame uses a different color range).
--xlim TEXT Set limits for the x-coordinate (lower,upper)
--ylim TEXT Set limits for the y-coordinate (lower,upper).
--legend / --no-legend Show legend.
--force-legend Force legend even when plotting a single
dataset.
-x, --xlabel TEXT Specify a x-axis label.
-y, --ylabel TEXT Specify a y-axis label.
--clabel TEXT Specify a label for colorbar.
--title TEXT Specify a title.
-i, --interval INTEGER Specify the animation interval.
--save Save figure as PNG.
--saveas TEXT Name to save the plot as.
--fps INTEGER Specify frames per second for saving.
--dpi INTEGER DPI (resolution) for output.
-e, --edgecolors TEXT Set color for cell edges (default: None)
--showgrid / --no-showgrid Show grid-lines (default: True)
--collected Animate a dataset that has been collected, i.e.
a single dataset with time taken to be the first
index.
--hashtag Turns on the pgkyl hashtag!
--show / --no-show Turn showing of the plot ON and OFF (default:
ON).
-h, --help Show this message and exit.
Suppose we extend the simulation time of the
gyrokinetic ion acoustic wave simulation
to tEnd=50 and the number of frames to nFrame=100, and run it
with
gkyl gk-ionSound-1x2v-p1.lua
Note
This simulation took 582 seconds running with decompCuts=8 (8 cores)
on a 2020 MacBookPro.
and plot the electrostatic potential from all frames with
pgkyl "gk-ionSound-1x2v-p1_phi_[0-9]*.bp" interp collect pl -x '$x$' -y '$\phi$' --group 1 --clabel 'time'
this produces the pretty picture below, showing the electrostatic potential as a function of \(x\) in each frame, with the color indicating the time stamp of that frame.
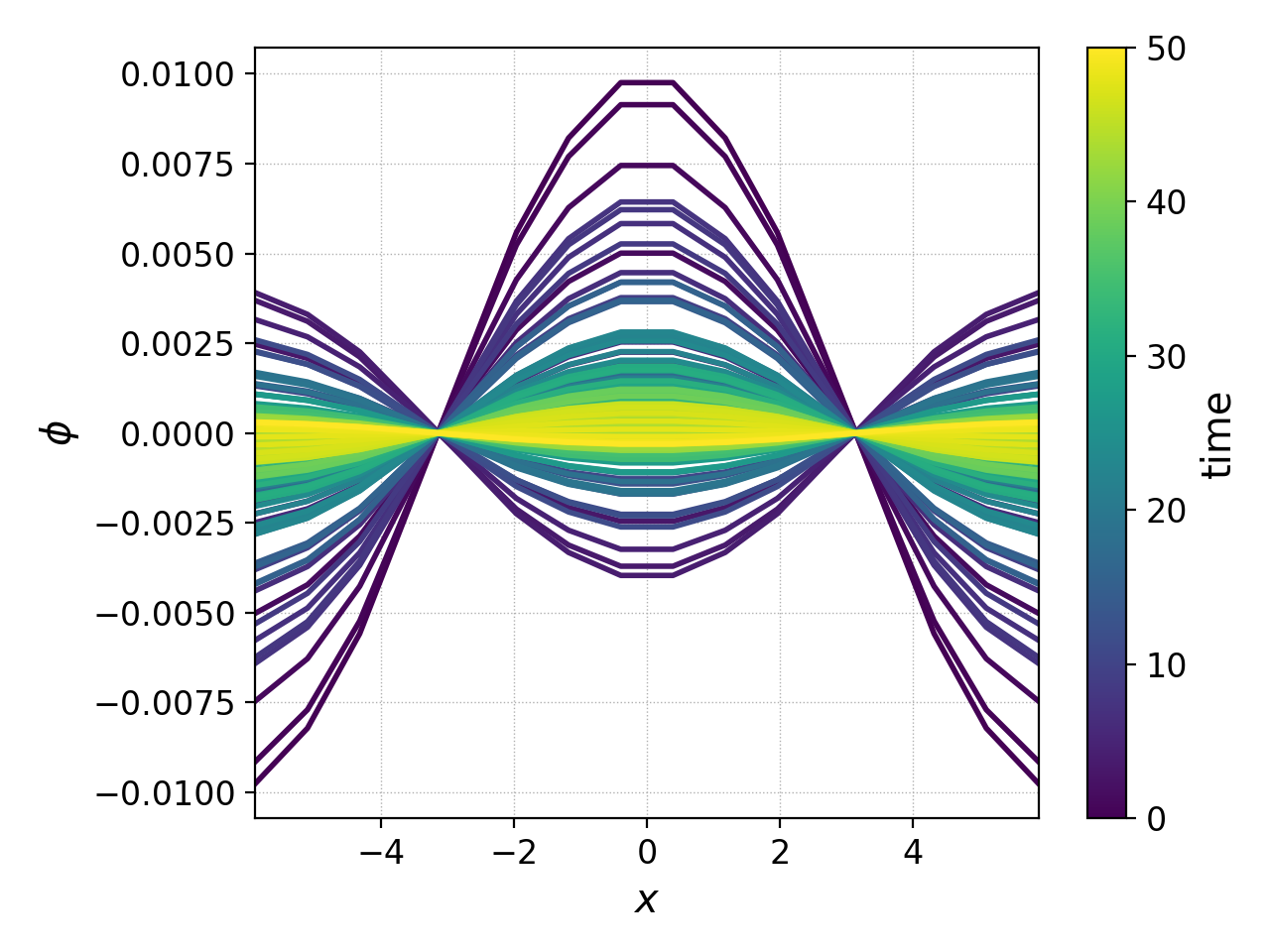
This kind of plot can become impractical if there are many frames, or if one would instead like to see a time-dependent movie of the evolution of the potential. For that purpose we can create an animation of \(\phi(x,t)\) using the following command
pgkyl "gk-ionSound-1x2v-p1_phi_[0-9]*.bp" interp anim -x '$x$' -y '$\phi$' --saveas 'gk-ionSound-1x2v-p1_phi.mp4'
Note
In order to save the animation to an .mp4 file, simply append
--saveas fileName.mp4 to the end of the above command.
and this will produce the animation below:
One can clearly see the amplitude of the wave decaying as Landau damping ruins the fun.