interpolate¶
Gkeyll’s simulation may employ higher-order methods with more than one degree of freedom per cell, typically expansion coefficients or nodal values. The interpolate command interpolates this higher-order representation onto a finer mesh with more points than the number of cells in the Gkeyll simulation.
Command line¶
Command help
pgkyl interpolate --help
Usage: pgkyl interp [OPTIONS]
Interpolate DG data onto a uniform mesh.
Options:
-b, --basistype [ms|ns|mo|mt] Specify DG basis.
-p, --polyorder INTEGER Specify polynomial order.
-i, --interp INTEGER Interpolation onto a general mesh of
specified amount.
-u, --use TEXT Specify a 'tag' to apply to (default all
tags).
-t, --tag TEXT Optional tag for the resulting array
-r, --read BOOLEAN Read from general interpolation file.
-n, --new for testing purposes
-h, --help Show this message and exit.
A Gkeyll simulation can have several degrees of freedom per cell. If we simply invoke the plot command we would then obtain a plot of all the coefficients in each cell. So for example, take the electron distribution function at the end of a two stream instability simulation and plot it with
pgkyl two-stream_elc_100.bp pl -x '$x$' -y '$v_x$'
we would obtain figure below on the left, with all 8 DG expansion
coefficients plotted in phase space. Most commonly we are interested
in plotting the actual function that the expansion coefficients
represent, rather than plotting such coefficients. We can do that
by interpolating onto a finer mesh with the interpolate command:
pgkyl two-stream_elc_100.bp interp pl -x '$x$' -y '$v_x$'
which results in the figure below on the right.
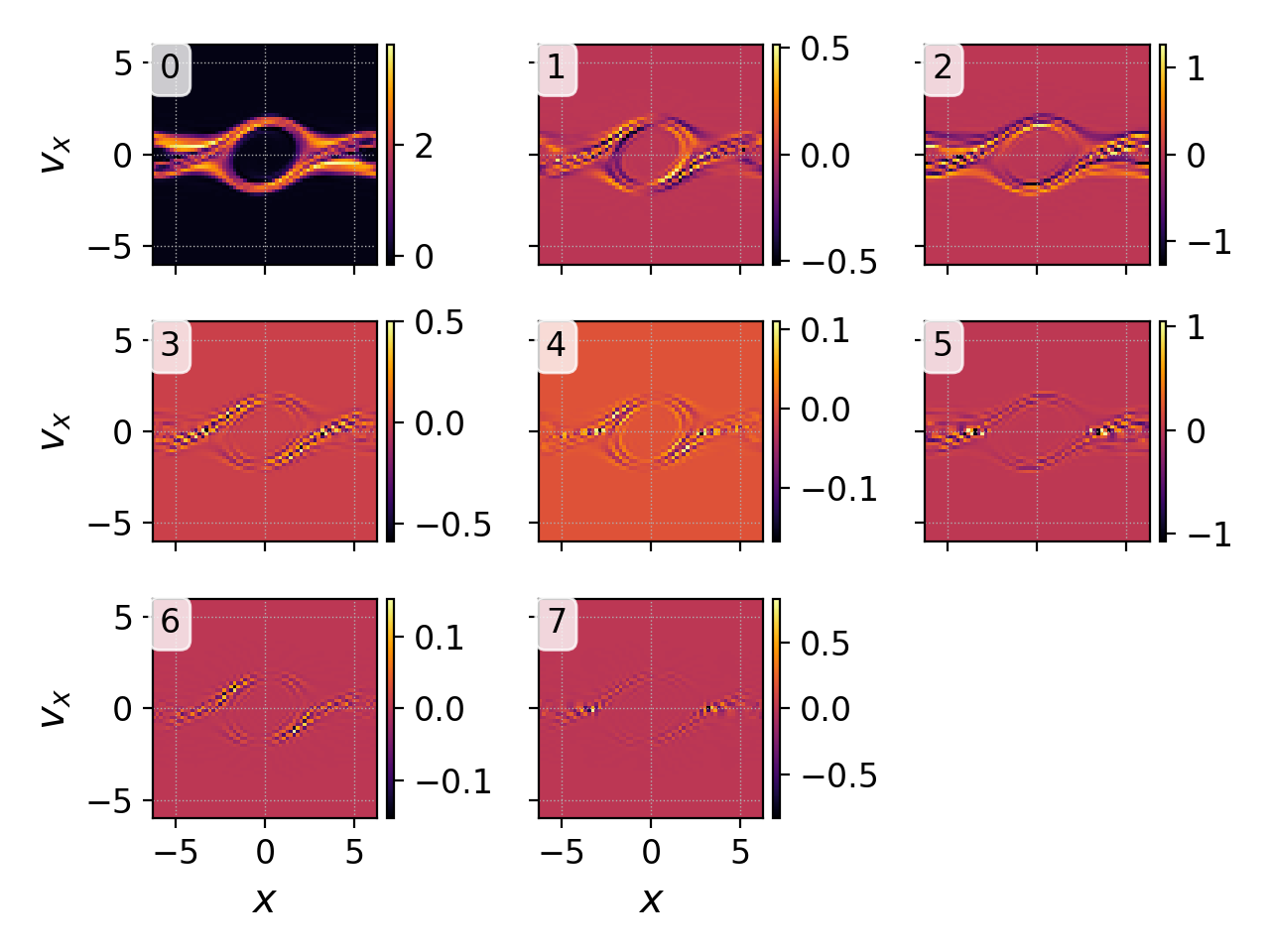
DG coefficients.¶
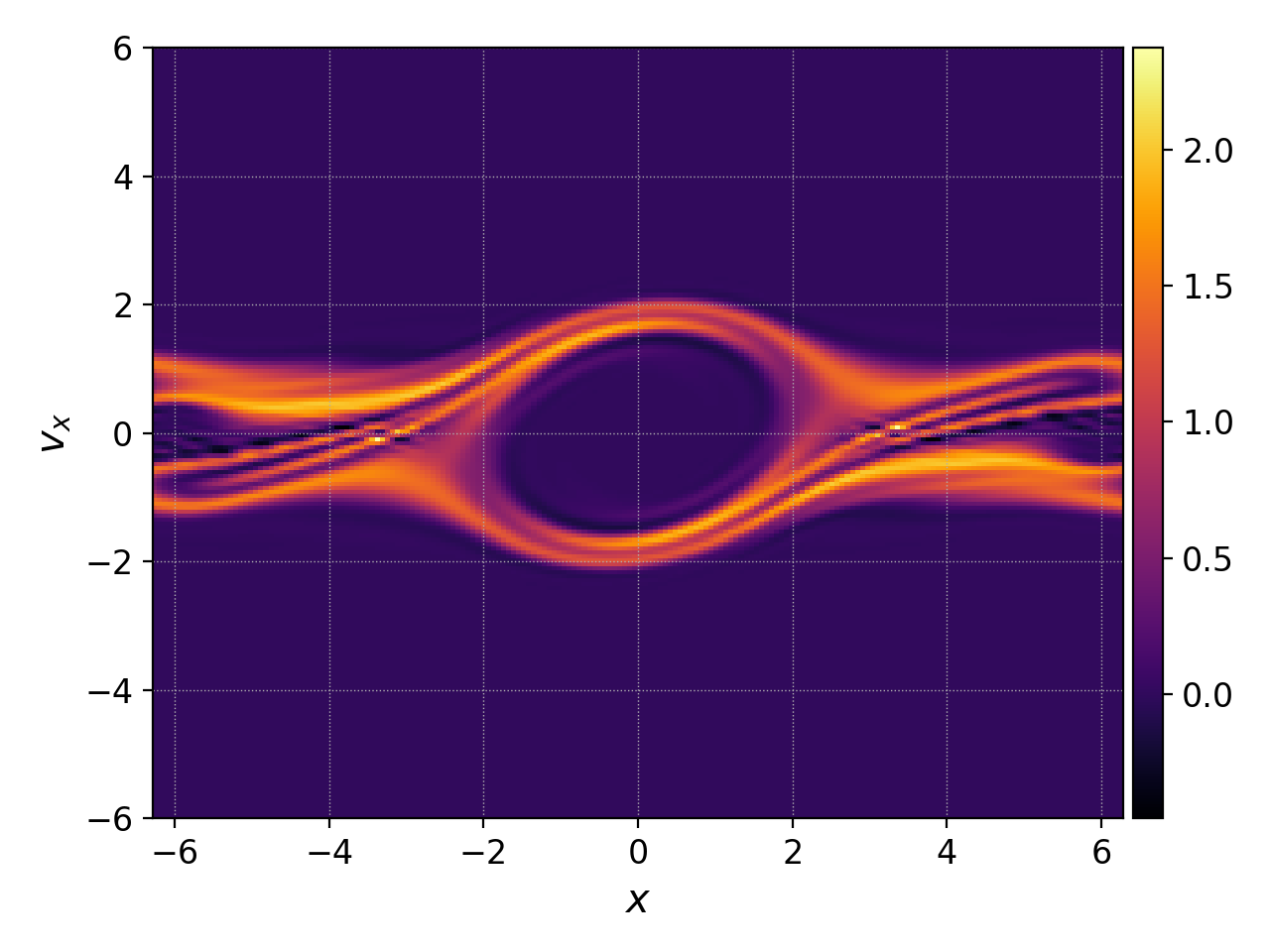
Interpolated distribution function.¶
We can compare the cell average of the electron distribution function with the interpolated distribution function with the following command
pgkyl two-stream_elc_100.bp -t fe interp -u fe -t fInterp sel -u fe -c0 -t c0 \
ev -l 'cell average' -t cellAv 'c0 2 /' activ -t fInterp,cellAv pl -b
which divides the zeroth DG coefficient by 2 in order to obtain the cell average (1r 2D piecewise quadratic basis), and produces the following figure
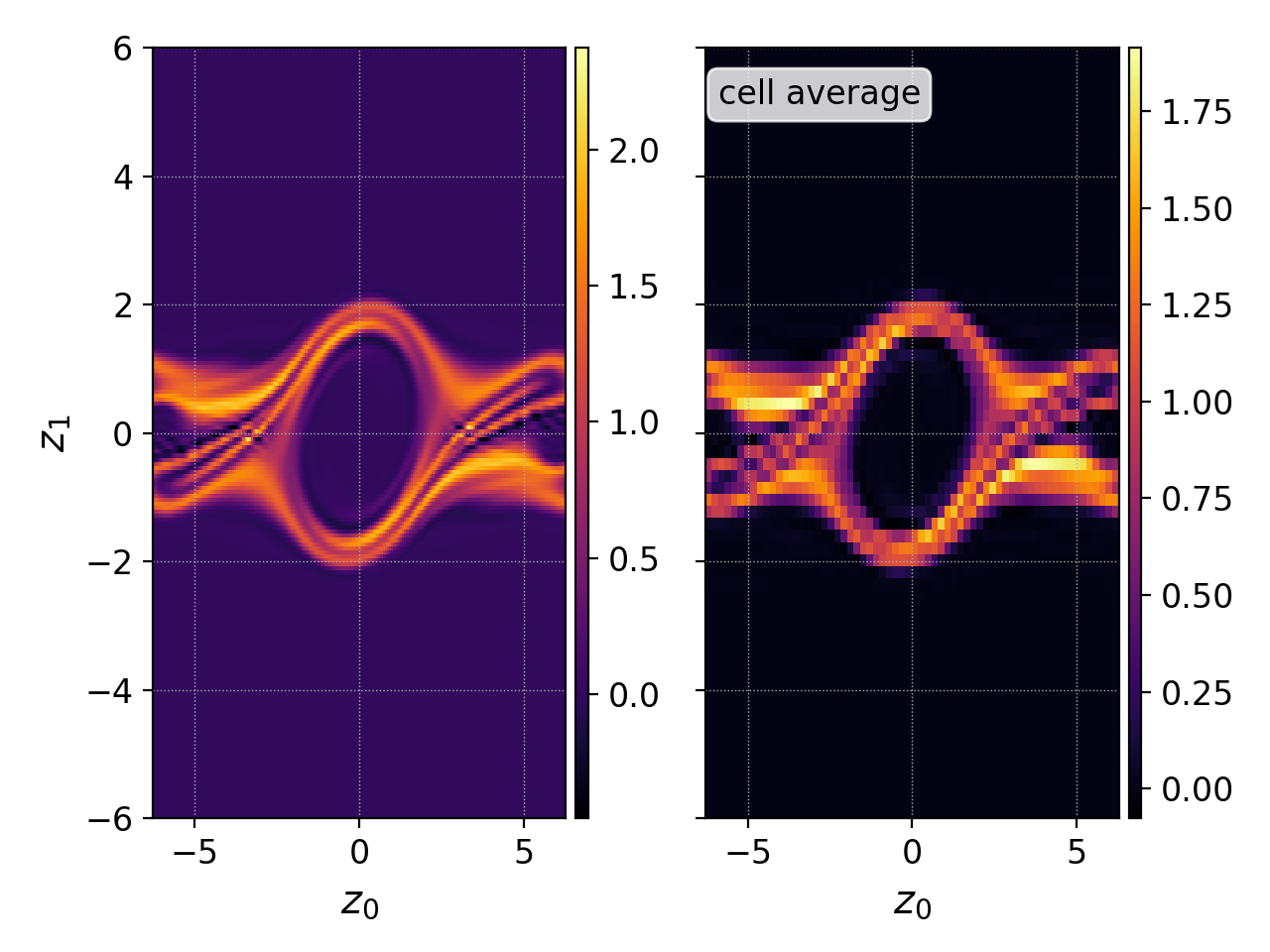
Notice how the cell average (right) is naturally coarser grained, and the interpolated function (left) offers a smoother plot.
By default the interpolate command interpolates onto a
uniform mesh that subdivides each cell in the simulation into
\(p+1\) cells in each direction, where \(p\) is the
polynomial order of the simulation. It is also possible to
interpolate onto finer meshes with the -i flag in order to
obtain smoother plots. However note that interpolating onto
finer meshes can also augment local maxima and/or minima. Below
we compare the final electron distribution function interpolated
onto a mesh with 3 subcells per cell in each direction (default
for \(p=2\)) and interpolating onto a mesh with 8 subcells
per cell in each direction:
pgkyl two-stream_elc_100.bp -t fe interp -t i3 interp -i 8 -u fe -t i8 activ -t i3,i8 pl -b -x '$x$' -y '$v_x$'
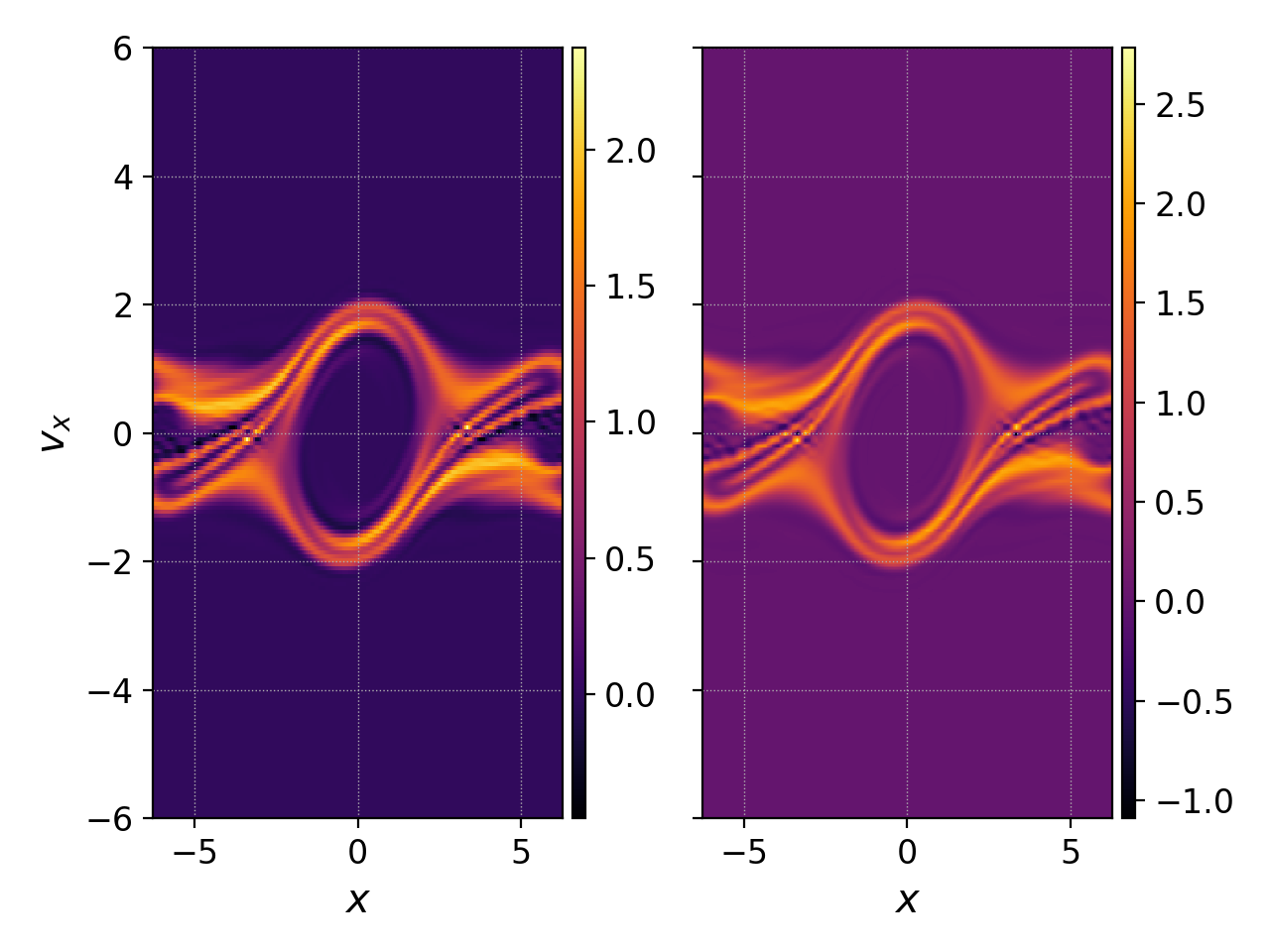
This example also shows the use of tags in order to tag datsets
and to instruct interpolate which datasets to operate on
(via the -u/--use flag). In order to request that
interpolate operates on a given tagged dataset, one must pass
-u followed by the dataset we wish to interpolate. And in
order to create a new dataset outof the interpolated data
one must use the -t flag followed by the name (tag) of the
new dataset. In the above example the first interpolate
operates on the input data (no -u necessary because it
immediately precedes it and there is only one dataset at that point
in the chain) and creates a dataset tagged i3. The second
interpolate operates on the input data (-u fe) and creates
a dataset tagged i8.
Script mode¶
interpolate uses the GInterpModal and GInterpNodal
classes based on the DG mode.
Parameter |
Description |
Default |
|---|---|---|
gdata (GData) |
A GData object to be used. |
|
polyOrder (int) |
The polynomial order of the discontinuous Galerkin discretization. |
|
basis (str) |
The polynomial basis. Currently supported options are |
After the initialization, both GInterpModal and GInterpNodal
can be used to interpolate data on a uniform grid and to calculate
derivatives
Member |
Description |
|---|---|
interpolate(int component, bool stack) -> narray, narray |
Interpolates the selected component (default is 0) of the DG data on a uniform grid |
derivative(int component, bool stack) -> narray, narray |
Calculates the derivative of the DG data |
When the stack parameter is set to true (it is false by
default), the grid and values are pushed to the GData stack rather
than returned.
An example of the usage:
import postgkyl as pg
data = pg.data.GData('bgk_neut_0.bp')
interp = pg.data.GInterpModal(data, 2, 'ms')
iGrid, iValues = interp.interpolate()