integrate¶
Integrate a dataset over specified direction(s).
Command line¶
Command help
$ pgkyl integrate --help
Usage: pgkyl integr [OPTIONS] AXIS
Integrate data over a specified axis or axes
Options:
-u, --use TEXT Specify the tag to integrate
-t, --tag TEXT Optional tag for the resulting array
-h, --help Show this message and exit.
Consider the gyrokinetic simulation of an ion acoustic wave as an example. It outputs the integrated particle density over time, which can plot as follows:
pgkyl gk-ionSound-1x2v-p1_ion_intM0.bp pl
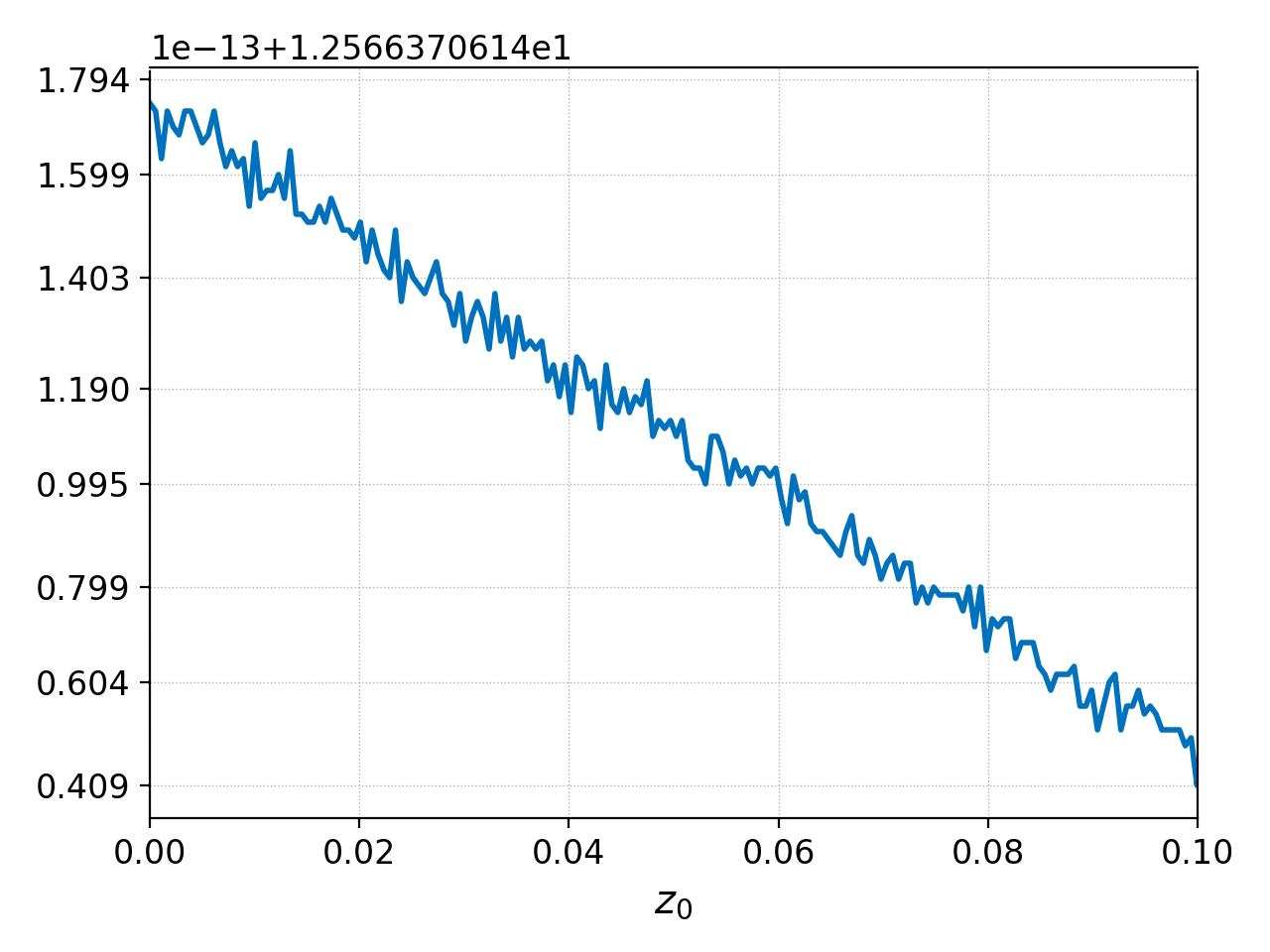
We can see from the values on the y-axis that the total number
of particles is 12.566. The number of particles should be conserved,
to machine precision. We can check this another way by integrating
the particle density along \(x\) (the 0th dimension) at the end
of the simulation with pgkyl:
pgkyl gk-ionSound-1x2v-p1_ion_GkM0_1.bp interp integr 0 print
where we have abbreviated integrate with integr, and we use
the print command to print the result of the integral to screen. The
output of this command is simply
12.566370614359034
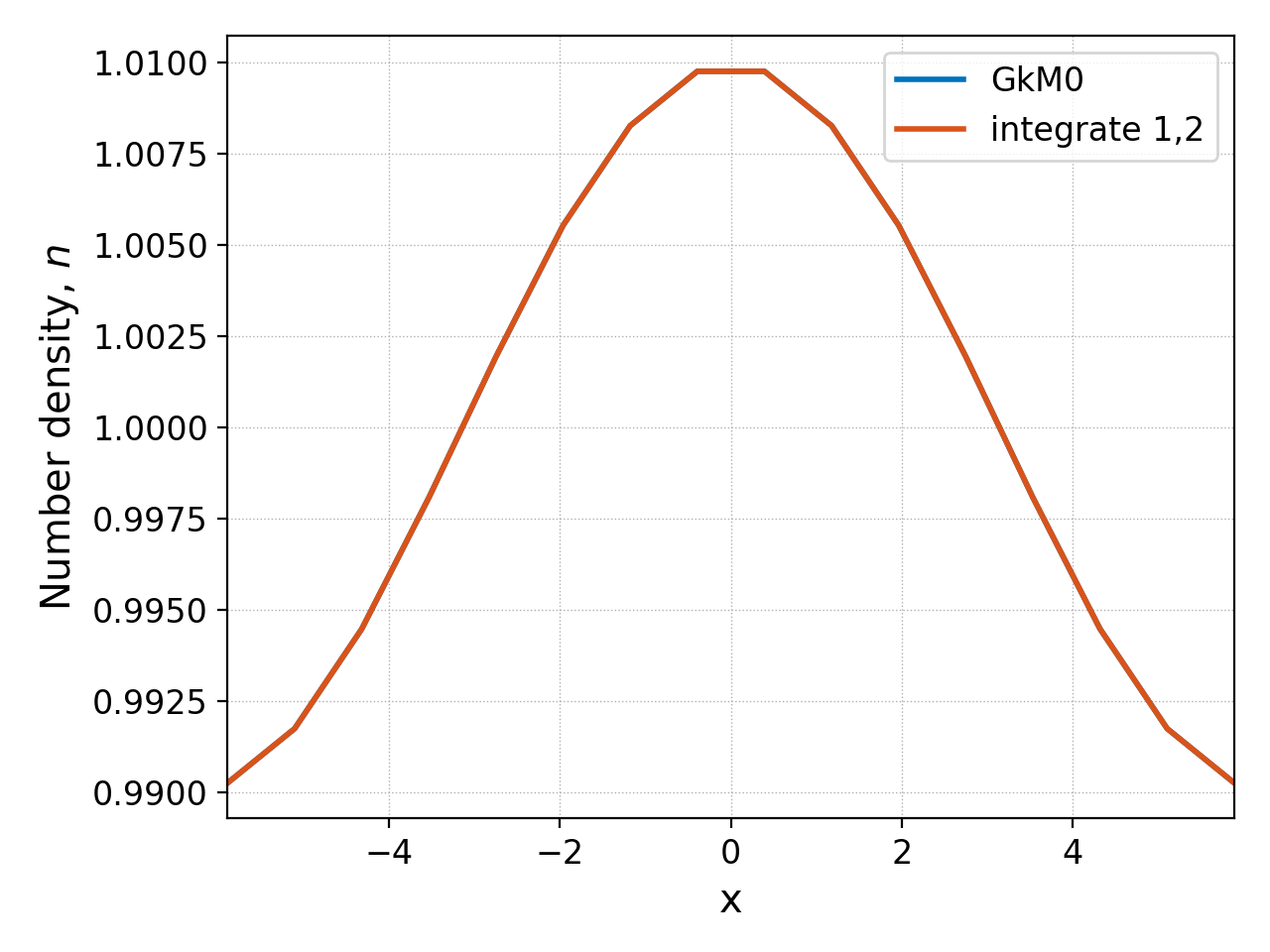
The integrate command can also be used to integrate higher dimensional datasets in one or more directions. We could take the ion distribution function and integrated along the \(v_\parallel\) and \(\mu\) directions (1st and 2nd dimensions, respectively) with
pgkyl gk-ionSound-1x2v-p1_ion_0.bp gk-ionSound-1x2v-p1_ion_GkM0_0.bp interp \
activate -i 0 integr 1,2 ev -l 'integrate 1,2' 'f[0] 6.283185 *' \
activate -i1,2 pl -f0 -x 'x' -y 'Number density, $n$'
Note
In this command we:
First load the ion distribution function (*_ion_0.bp) and its number density (*_ion_GkM0_0.bp) at \(t=0\).
Integrate the distribution function over velocity space with
activate -i 0 integr 1,2.Multiply such integral by \(2\pi B_0/m_i\) (\(B_0=m_i=1\) here) with
ev -l 'integrate 1,2' 'f[0] 6.283185 *'.Activate the number denstiy and integrated distribution function data sets and plot them with
activate -i1,2 pl -f0.
and this should give approximately the same number density as the
GkM0 diagnostic outputted by the simulation, as shown below.
Another useful application of the integrate command is to integrate,
or average, over time (although note that the ev
command has a avg operation that may make this easier). Usually
this requires collecting multiple frames into a single dataset with
the collect command, and then integrating over the
0th dimension (time).
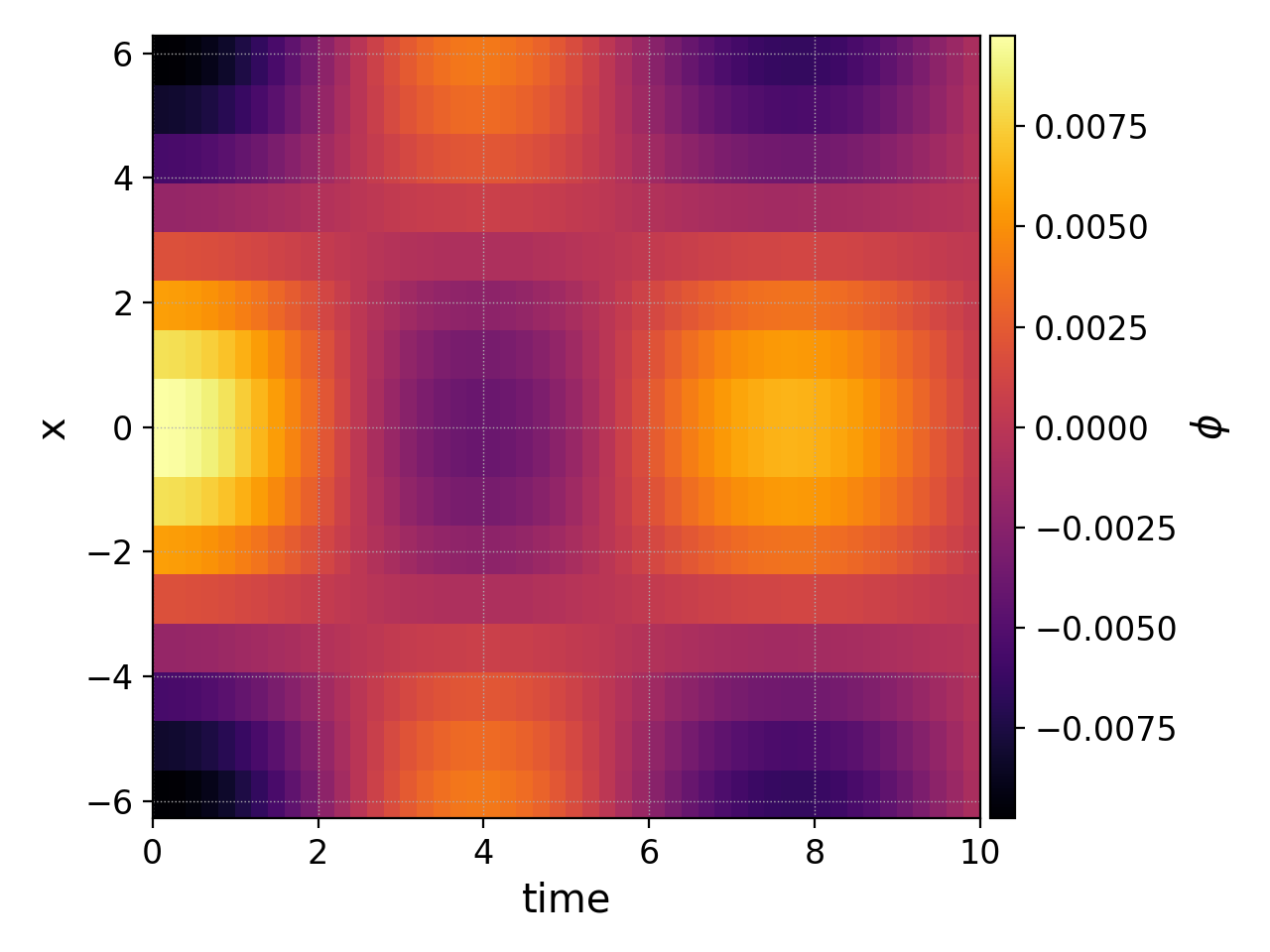
So if we increase the tEnd of the gyrokinetic ion sound wave
simulation to 10 and the number of frames to 50 we could
plot the electrostatic potential as a function of time and position
with
pgkyl "gk-ionSound-1x2v-p1_phi_[0-9]*.bp" interp collect pl -x 'time' -y 'x' --clabel '$\phi$'
We can integrate this potential in time and plot it on top of the initial potential with
pgkyl gk-ionSound-1x2v-p1_phi_0.bp -l '$t=0$' -t phi0 \
"gk-ionSound-1x2v-p1_phi_[0-9]*.bp" -t phis interp collect -u phis -t phiC \
integrate -u phiC -t phiInt 0 ev -l 'Time average' -t phiAvg 'phiInt 10. /' \
activate -t phi0,phiAvg pl -f0 -x '$x$' -y '$\phi$'
This command uses tags to select which dataset to perform an operation on. The end result is the plot below
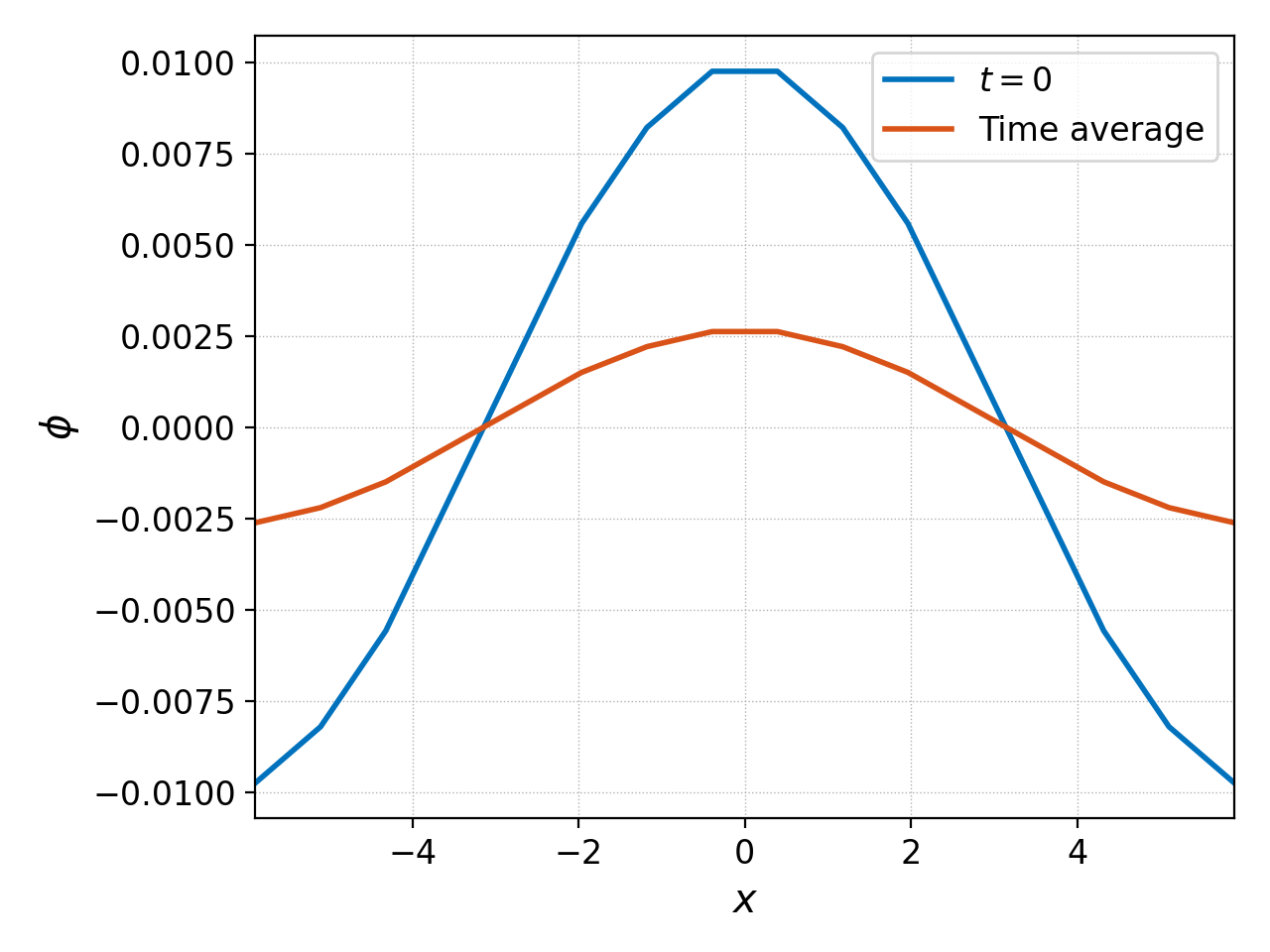
showing that the time averaged potential is lower amplitude due to the collisionless Landau damping of the wave.